Zusammenfassung
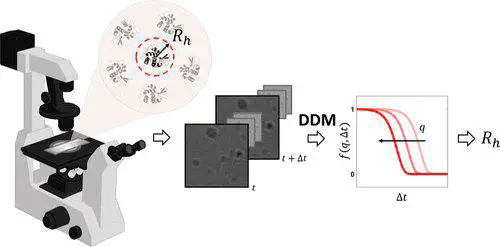
Introduced more than 50 years ago, dynamic light scattering (DLS) is routinely used to determine the size distribution of colloidal suspensions as well as of macromolecules in solution, such as proteins, nucleic acids, and their complexes. More recently, differential dynamic microscopy (DDM) has been proposed as a way to perform DLS experiments with a microscope, with much less stringent constraints in terms of cleanliness of the optical surfaces but a potentially lower sensitivity due to the use of camera-based detectors. In this work, we push bright-field DDM beyond known limits and show it to be sufficiently sensitive to size small macromolecules in diluted solutions. By considering solutions of three different proteins (bovine serum albumin, lysozyme, and pepsin), we accurately determine the diffusion coefficient and hydrodynamic radius of both single proteins and small protein aggregates down to concentrations of a few milligrams per milliliter. In addition, we present preliminary results showing an unexplored potential for the determination of virial coefficients. Our results are in excellent agreement with those obtained in parallel with a state-of-the-art commercial DLS setup, showing that DDM represents a valuable alternative for rapid, label-free protein sizing with an optical microscope.
Add the publication’s full text or supplementary notes here. You can use rich formatting such as including code, math, and images.